

After identifying the experimental unit and the number of replications that will be used, the next step is to assign the treatments (i.e. factor levels or factor level combinations) to experimental units. In a completely randomized design, treatments are assigned to experimental units at random. This is typically done by listing the treatments and assigning a random number to each. In the greenhouse experiment discussed in Chapter 1, there was a single factor (fertilizer) with 4 levels (i.e. 4 treatments), six replications, and a total of 24 experimental units (each unit a potted plant). Suppose the image below is the Greenhouse Floor plan and bench that was used for the experiment (as viewed from above). .png?revision=1) We need to be able to randomly assign each of the treatment levels to 6 potted plants. To do this, assign physical position numbers on the bench for placing the pots.
We need to be able to randomly assign each of the treatment levels to 6 potted plants. To do this, assign physical position numbers on the bench for placing the pots. .png?revision=1)
Steps in Minitab In Minitab, this assignment can be done by manually creating two columns: one with each treatment level repeated 6 times (order not important) and the other with a position number 1 to \(N\), where \(N\) is the total number of experimental units to be used (i.e. \(N=24\) in this example). The third column will store the treatment assignment. 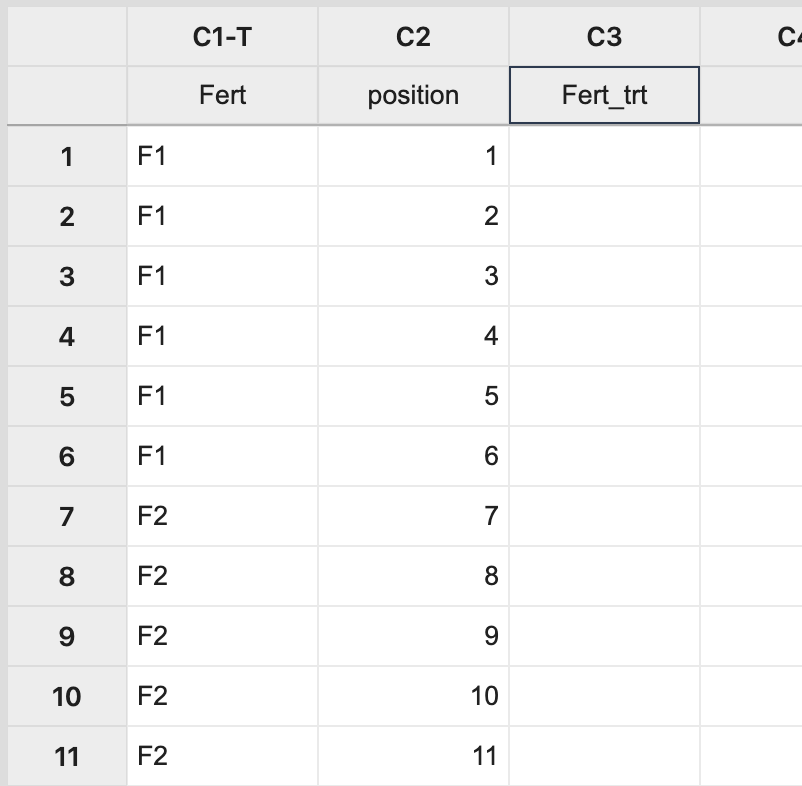 Next, select Calc > Sample from Columns, fill in the dialog box as seen below, and click OK.
Next, select Calc > Sample from Columns, fill in the dialog box as seen below, and click OK. 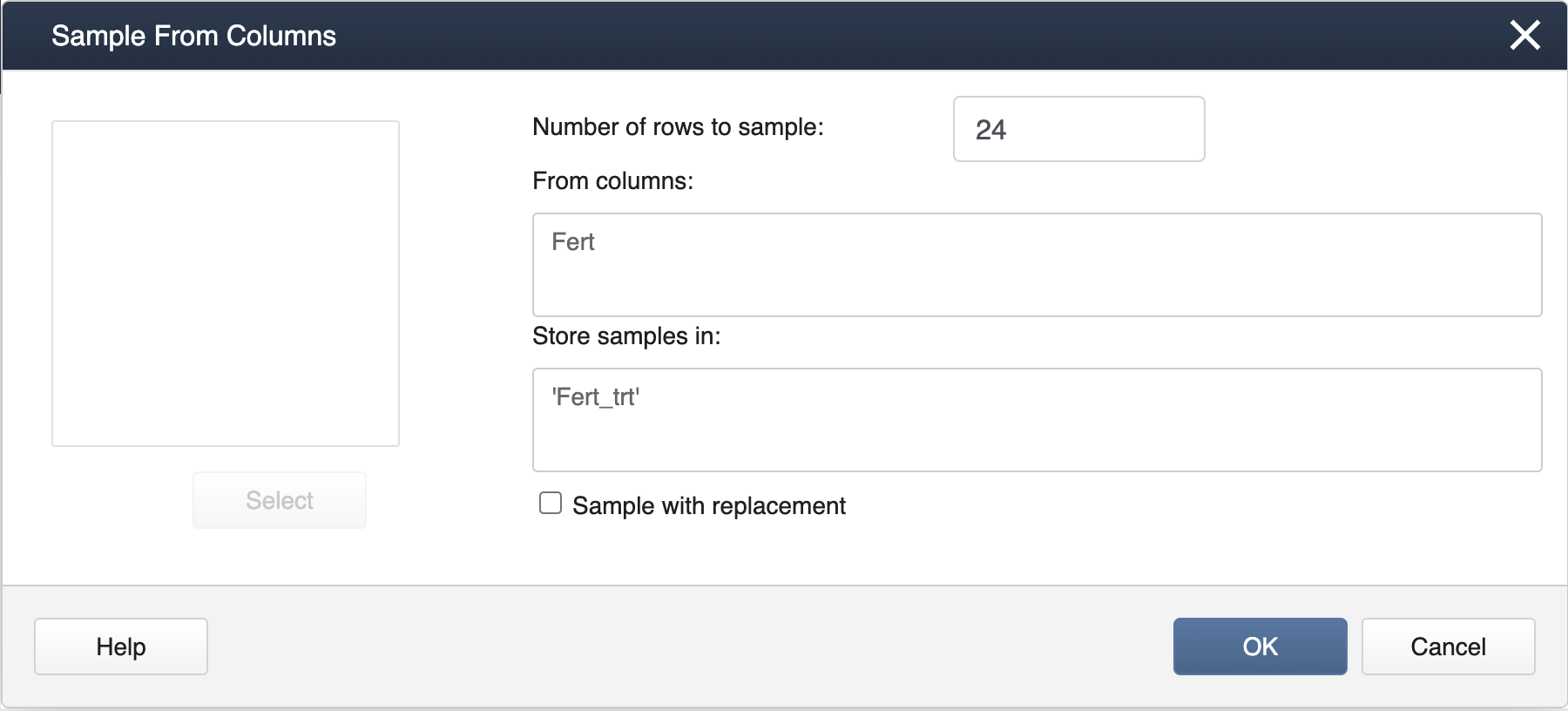
Be sure to have the "Sample with Replacement" box unchecked so that all treatment levels will be assigned to the same number of pots, giving rise to a proper completely randomized design for a specified number of replicates.
This will result in a completely random assignment. 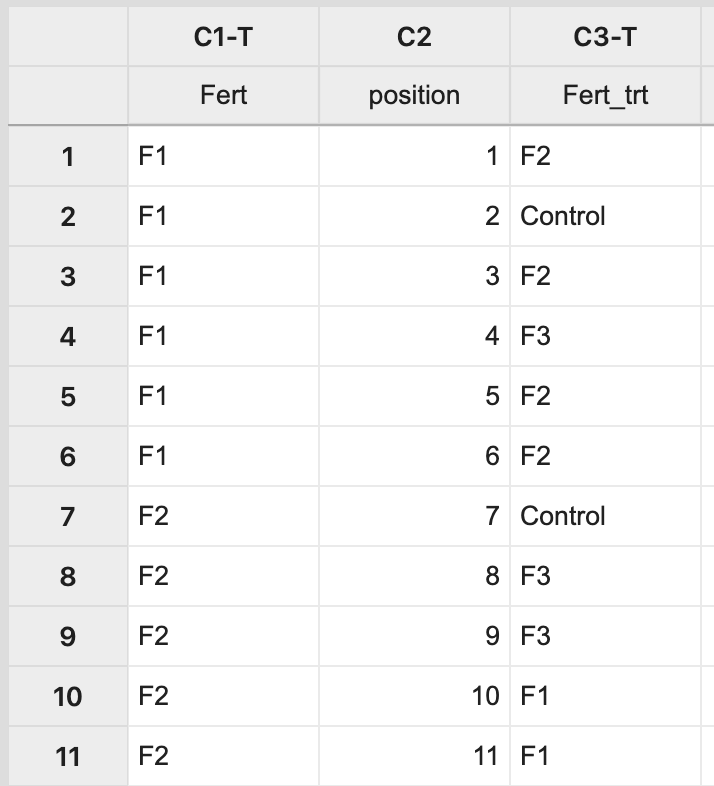 This assignment can then be used to apply the treatment levels appropriately to pots on the greenhouse bench.
This assignment can then be used to apply the treatment levels appropriately to pots on the greenhouse bench. .png?revision=1)
proc surveyselect data=greenhouse out=trtassignment outrandom method=srs samprate=1; run;
The output would be as below. In practice, it is recommended to specify a seed to ensure the results are reproducible.
| Obs | Fertilizer |
|---|---|
| 1 | F3 |
| 2 | F2 |
| 3 | Con |
| 4 | F2 |
| 5 | F3 |
| 6 | Con |
| 7 | F2 |
| 8 | F2 |
| 9 | F3 |
| 10 | F1 |
| 11 | F1 |
| 12 | F3 |
| 13 | F2 |
| 14 | F1 |
| 15 | F3 |
| 16 | F3 |
| 17 | F1 |
| 18 | Con |
| 19 | Con |
| 20 | F2 |
| 21 | Con |
| 22 | F1 |
| 23 | Con |
| 24 | F1 |
sample(treatment) [1] "F3" "F2" "F1" "F2" "F3" "F1" "Control" "F2" "F3" [10] "F3" "F2" "Control" "F3" "F1" "F1" "F2" "Control" "F2" [19] "F1" "Control" "F3" "Control" "Control" "F1"
This means that the first experimental unit will get Fertilizer 3, the second experimental unit will get Fertilizer 2, etc.
Obtain the block design. Load the greenhouse data and obtain the ANOVA table. To obtain the block design we can use the following commands:
library(blocksdesign) block_design# obs block plant treatment # 1 1 1 1 4 # 2 2 1 2 1 # 3 3 1 3 3 # 4 4 1 4 2 # 5 5 2 1 1 # 6 6 2 2 4 # 7 7 2 3 3 # 8 8 2 4 2 # 9 9 3 1 3 # 10 10 3 2 1 # 11 11 3 3 4 # 12 12 3 4 2 # 13 13 4 1 1 # 14 14 4 2 4 # 15 15 4 3 2 # 16 16 4 4 3 # 17 17 5 1 3 # 18 18 5 2 2 # 19 19 5 3 1 # 20 20 5 4 4 # 21 21 6 1 2 # 22 22 6 2 1 # 23 23 6 3 4 # 24 24 6 4 3
To load the greenhouse data and obtain the ANOVA table ( lmer() and aov( )) we use the following commands:
setwd("~/path-to-folder/") greenhouse_RCBD_data read.table("greenhouse_RCBD_data.txt",header=T) attach(greenhouse_RCBD_data) library(lmerTest) library(lme4) greenhouse_RCBD_anova<-lmer(Height ~ Fertilizer + (1 | factor(Block)),greenhouse_RCBD_data) anova(greenhouse_RCBD_anova) #Type III Analysis of Variance Table with Satterthwaites method # Sum Sq Mean Sq NumDF DenDF F value Pr(>F) #Fertilizer 251.44 83.813 3 15 162.96 1.144e-11 *** #--- #Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 greenhouse_RCBD_anova1#Error: factor(Block) # Df Sum Sq Mean Sq F value Pr(>F) #Residuals 5 53.32 10.66 #Error: Within # Df Sum Sq Mean Sq F value Pr(>F) #Fertilizer 3 251.44 83.81 163 1.14e-11 *** #Residuals 15 7.72 0.51 #--- #Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
For comparison the ANOVA table for the completely randomized design is given below:
greenhouse_CRD_anova# Df Sum Sq Mean Sq F value Pr(>F) #Fertilizer 3 251.44 83.81 27.46 2.71e-07 *** #Residuals 20 61.03 3.05 #--- #Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 detach(greenhouse_RCBD_data)
This page titled 7.2: Completely Randomized Design is shared under a CC BY-NC 4.0 license and was authored, remixed, and/or curated by Penn State's Department of Statistics.